About RESIST
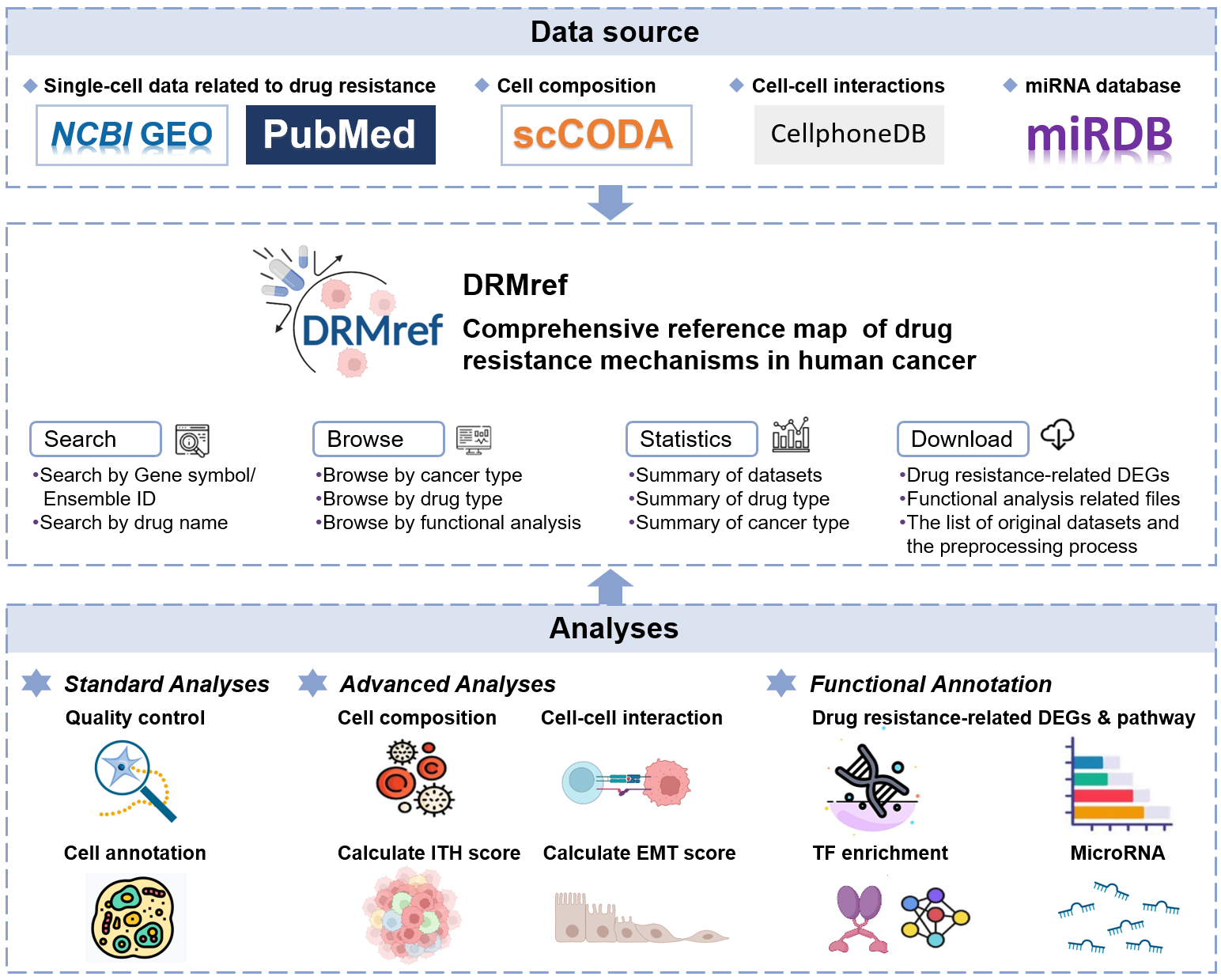
Drug resistance is a pervasive challenge in cancer treatment. The emergence and development of single-cell technology enable us to investigate the mechanisms of drug resistance at the individual cell and cell type levels. Therefore, we established the DRMref database, which aims to provide comprehensive characterization of drug resistance mechanisms using single-cell data obtained from drug treatment settings.
The current version of DRMref database includes 42 scRNA datasets from 30 studies, 14 of which have both pre- and post-treatment samples, covering 666 samples, 13 major cancer types, 26 minor cancer types, 35 treatment regimens, and 42 drugs.
We conducted analysis on the differences in cell composition, intra-tumor heterogeneity (ITH) and epithelial-mesenchymal transition (EMT) scores, cell-cell interactions, and differentially expressed genes between the resistant and sensitive groups. DRMref also performed enrichment analysis of 6 known drug resistance mechanisms, Hallmark, KEGG and GO BP pathways. Additionally, We provide microRNAs, motifs and transcription factors (TFs) realted to drug resistance-related genes. DRMref will serve as a unique resource for studing drug resistance, drug combination therapy and discovering new targets.
Search & Browse Options
Explore our database with multiple search options for genes, drugs, diseases, and functional analysis.
Gene Search
Search by gene symbol or Ensembl ID
Drug Search
Search by drug names or browse by types
Disease Browse
Browse by cancer types and explore results
Functional Analysis
Explore DEGs, pathways, and regulatory elements
Analysis Your Customized Data
Upload your own data and generate the same comprehensive 15-section visualization analysis results as our existing dataset pages. Our analysis pipeline supports various data types and provides in-depth insights into drug resistance mechanisms.
Supported Data Types
- • Single-cell RNA sequencing
- • Bulk RNA sequencing
- • RNA sequencing
- • Gene microarray
Analysis Features
- • Differential expression analysis
- • Functional enrichment analysis
- • ITH & EMT analysis
- • miRNA & RBP analysis
Copyright 2025-Present - University of Florida
